How bacteria unify to protect against antibiotics
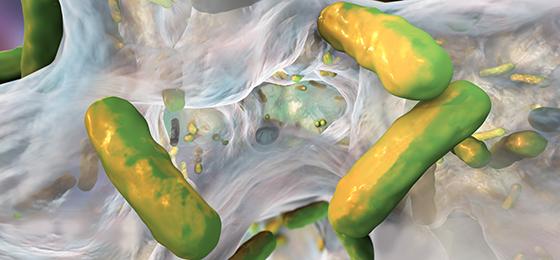
Bacteria in biofilms are difficult to target with antibiotics. New findings could pave the way for new treatment strategies.
Project description (completed research project)
Biofilms protect bacteria against antibiotics
Most bacterial infections are caused by biofilms. These are bacterial cells together with their self-produced matrix that form a dense coating to occupy a range of surfaces including those of medical products such as implants. In a biofilm, the bacteria surround themselves with protective gelatinous substances comprised of extracellular components. As a result, when treating infections with antibiotics only a fraction of the active agent is able to come into contact with the bacteria, leading to sub-lethal concentrations of antibiotics and thus helping to generate resistance. In the international research project BEAT-AMR, with partners from Switzerland, Germany, the UK and the Netherlands, Qun Ren and colleagues at Empa investigated how bacteria in biofilms adapt to antibiotics, how they acquire resistance and spread them, and how this influences the bacterial composition in the biofilm. There has been no previous systematic research conducted on the influence of biofilms on the formation of resistance.
The team at Empa focused on the identification and characterisation of genes involved in biofilm-specific resistance of Pseudomonas aeruginosa towards antibiotics. P. aeruginosa is a pathogen of high clinical relevance, responsible for many deadly infections for humans and showing high recalcitrance to current antibiotic treatments. Within biofilms, P. aeruginosa cells can resist higher antibiotic concentration than what is allowed in the human body. Thus, new therapeutic approaches are urgently needed to improve P. aeruginosa removal in the context of biofilm-related infections.
Several genes involved in antibiotic tolerance identified
Qun Ren and her team first established and validated a model system to identify genes involved in biofilm growth and antibiotic resistance. They characterised the important bacterial strain P. aeruginosa MPAO1 in much detail, at the genomic level (= totality of genetic information of an organism), proteomic level (= totality of all proteins in an organism), and phenotypic level ( = observable physical properties of an organism). They discovered several key genetic determinants responsible for making P. aeruginosa biofilms tolerant. Tolerant bacteria are not antibiotic-resistant, but they can temporarily fall into a sleep-like state and thus escape the effect of antibiotics. In this way they persist in patients, often leading to chronic infections that are difficult to treat. Furthermore, tolerance is thought to be a precursor to resistance.
The identification of several sets of genes involved in biofilm-related P. aeruginosa tolerance provided important novel insights, which can help to predict the evolution of tolerance in patients and facilitate the development of targeted treatment strategies. Furthermore, a number of genes showed high clinical potential as targets for new antibiotics.
November 2021
Original title
Partnership against Biofilm-associated Expression, Acquisition and Transmission of AMR
