Nanosensors significantly accelerate diagnosis
Researchers at the University of Basel have developed a method that greatly accelerates the identification of antibiotic-resistant pathogens.
Project description (completed research project)
In any treatment of infections with multi-resistant bacteria, accurate identification of the pathogens and resistance types is vital. Traditional methods that are currently used routinely for this diagnosis require relatively large quantities of bacteria, which are multiplied from samples taken from patients. Depending on the pathogen, it can take up to 72 hours before a sufficient quantity of bacteria is available for the diagnosis. Because of this, valuable time that could have been used for treatment is lost. And doctors often resort to using antibiotics haphazardly before clear results are obtained. But this practice promotes the development of resistance.
Sensors bind resistance genes
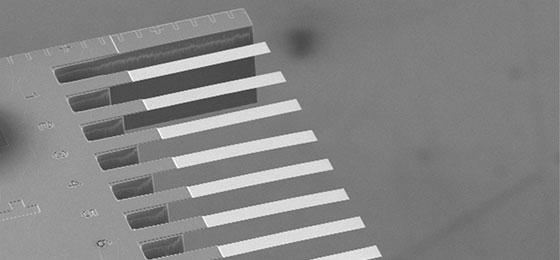
Ernst Meyer and his team at the University of Basel have now developed a much faster technique employing a completely new technology in this field that uses microscopically small sensors (nanomechanical cantilevers) coated with various biomarkers. These markers precisely match the shape to which individual and very specific genetic sequences of a bacterium bind. As a result, they can be made to specifically bind those sequences that are responsible for various types of resistance. When a bacterial sample containing the relevant sequence comes into contact with a nanosensor, the surface tension of the sensor is measurably changed. This change shows whether a pathogen possesses a specific resistance.
Fast and reliable results
The great advantage of this method is that the bacteria do not need to be multiplied in order to obtain reliable results. Rather, the RNA samples isolated directly from bacteria are sufficient. The researchers in Ernst Meyer's team have now developed sensors that bind various frequently occurring vancomycin resistance genes. In a comprehensive series of tests, their method detected these resistances just as reliably as the existing standard tests, but with a processing time from sample collection to the result of under an hour. This significantly accelerates the diagnostic process.
Comprehensive resistance tests possible
However, the developed prototype is not yet ready for use in practice, since it can be operated only with considerable technical expertise. But given the impressive accuracy and speed of the method, as well as the comparatively cheap materials involved, it will now be further developed to make it easier to operate. Meyer and his team are also expanding its use to other types of resistance. With several serially connected sensors, comprehensive resistance tests can be carried out very quickly. The next step for the researchers is to focus on diagnosis in cases involving sepsis, since the rapid detection of antibiotic resistance is particularly important in such situations and can mean the difference between life and death.
February 2022
Original title
Fast Assessment of antibiotic resistance in bacteria by using nanomechanical arrays